Converting Amazon Textract tables to pandas DataFrames
I’m currently doing a lot of document processing at work. One of my tasks is to extract tables from PDF files. I evaluated Amazon Textract’s table extraction capability as part of this task. It’s very well documented, as is the rest of Textract. I was slightly disappointed by the examples, but nothing serious.
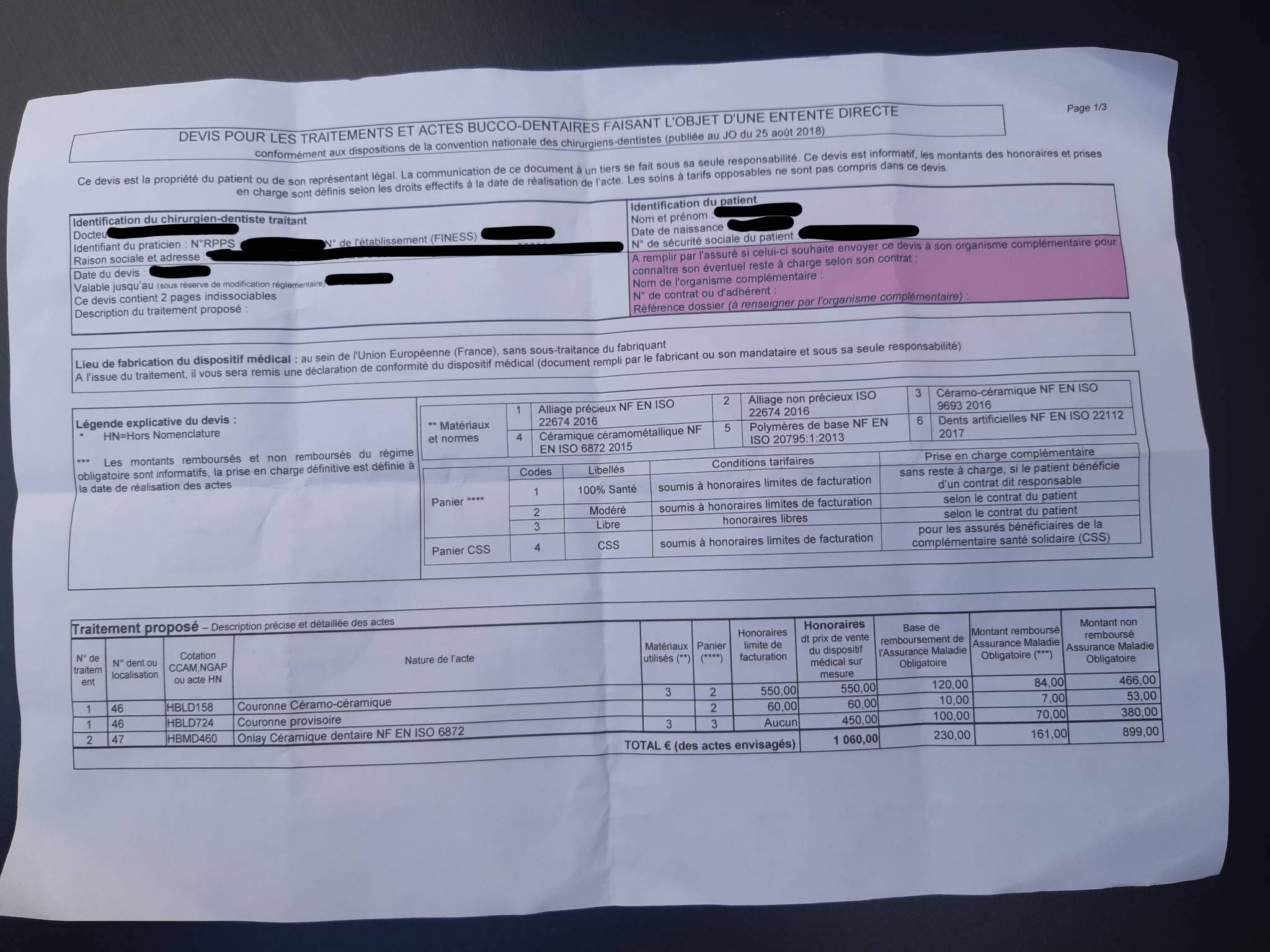
I wanted to write this short blog post to share a piece of code I use to convert tables extracted through Amazon Textract to pandas.DataFrames. I’ll be using the following anonymised image as an example:
You can process PDF files with Amazon Textract, but only in asynchronous mode. That implies uploading a job to a queue and waiting for the job to be done. You can however process images in synchronous mode via a simple API call. Therefore, you could convert a PDF file to an image in Python with pdf2image in order to process it synchronously. In this example I’m just using an image though.
The first step is to load the image, which can be done with PIL:
from PIL import Image
im = Image.open('example.jpg')
Amazon Textract expects the image to be encoded via the Base64 encoding scheme. This is very easy to do in Python:
import io
buffered = io.BytesIO()
im.save(buffered, format='PNG')
Next, we want to call the Amazon Textract API. The easiest way to proceed is to use boto3, which is the official Python SDK for interacting with AWS. Setting up boto3 and linking it to your AWS account is well explained in the official documentation. Once this is done, calling Textract is trivial:
import boto3
client = boto3.client('textract')
response = client.analyze_document(
Document={'Bytes': buffered.getvalue()},
FeatureTypes=['TABLES']
)
We specify the 'TABLES' feature to tell Textract to find the tables in the document. There’s also a 'FORMS' feature, but that’s for key/value forms, where the keys and the values are typically separated by two points (:).
The response from Textract is made up of a list of blocks. Each block represents a piece of the document. A block is defined by a geometric bounding box. Blocks are related to each other in a parent/child paradigm. There are four types of blocks which we are interested in:
- A
WORD. - A
SELECTION_ELEMENT, which is essentially a checkbox. - A
CELL, which is a block that contains one or moreWORDs and/or aSELECTION_ELEMENT. - A
TABlE, which is made up ofCELLs.
The response data is not nested. The children of a block are specified in said block. For instance, here’s a CELL block:
{'BlockType': 'CELL',
'ColumnIndex': 1,
'ColumnSpan': 1,
'Confidence': 99.99826049804688,
'Geometry': {'BoundingBox': {'Height': 0.06898262351751328,
'Left': 0.055694881826639175,
'Top': 0.6720325350761414,
'Width': 0.02915901131927967},
'Polygon': [{'X': 0.055994194000959396, 'Y': 0.6733753085136414},
{'X': 0.084853895008564, 'Y': 0.6720325350761414},
{'X': 0.08466880023479462, 'Y': 0.7396966218948364},
{'X': 0.055694881826639175,
'Y': 0.7410151362419128}]},
'Id': '2bbfde52-e68a-4d5d-b479-2a86d8525dbb',
'Relationships': [{'Ids': ['92aec878-0863-4acc-afd5-3eb6515adc93',
'199d0652-0e90-43ac-bc74-b28d2fad35c7',
'383fb964-2da1-4bb5-824a-5978b24d790a',
'8bb647da-46ed-4947-8caa-568960569d7e'],
'Type': 'CHILD'}],
'RowIndex': 1,
'RowSpan': 1}
The goal is to munge this data and convert the list of blocks to a pandas.DataFrame. I first mapped each ID to the corresponding block. I did this for the four kind of blocks we may encounter.
def map_blocks(blocks, block_type):
return {
block['Id']: block
for block in blocks
if block['BlockType'] == block_type
}
blocks = response['Blocks']
tables = map_blocks(blocks, 'TABLE')
cells = map_blocks(blocks, 'CELL')
words = map_blocks(blocks, 'WORD')
selections = map_blocks(blocks, 'SELECTION_ELEMENT')
I then wrote a little function that yields the children IDs of a block.
def get_children_ids(block):
for rels in block.get('Relationships', []):
if rels['Type'] == 'CHILD':
yield from rels['Ids']
The dataframe construction is logic is rather straightforward that we’ve done the ID/block mapping. Obviously we’re going to loop over each table. Then we’re going to list the cells which belong to the table. We can then determine the number of cells and rows by looking at the RowIndex and ColumnIndex fields of each cell. Next, we loop over each cell and retrieve the cell’s content, which can be either be words or checkboxes. We store the cell contents in a list of lists that represents the table. Once we’re done looping over the cells, we can instantiate the pandas.DataFrame by assuming that the first row represents the column headers.
import pandas as pd
dataframes = []
for table in tables.values():
# Determine all the cells that belong to this table
table_cells = [cells[cell_id] for cell_id in get_children_ids(table)]
# Determine the table's number of rows and columns
n_rows = max(cell['RowIndex'] for cell in table_cells)
n_cols = max(cell['ColumnIndex'] for cell in table_cells)
content = [[None for _ in range(n_cols)] for _ in range(n_rows)]
# Fill in each cell
for cell in table_cells:
cell_contents = [
words[child_id]['Text']
if child_id in words
else selections[child_id]['SelectionStatus']
for child_id in get_children_ids(cell)
]
i = cell['RowIndex'] - 1
j = cell['ColumnIndex'] - 1
content[i][j] = ' '.join(cell_contents)
# We assume that the first row corresponds to the column names
dataframe = pd.DataFrame(content[1:], columns=content[0])
dataframes.append(dataframe)
In our case we have two tables.
>>> len(dataframes)
2
The first table is the one I’m interested in extracting.
| N° de traitem ent | N° dent ou localisation | Cotation CCAM,NGAP ou acte HN | Nature de l’acte | Matériaux utilisés (**) | Panier (****) | Honoraires limite de facturation | Honoraires dt prix de vente du dispositif médical sur mesure | Base de remboursement de l’Assurance Maladie Obligatoire | Montant remboursé Assurance Maladie Obligatoire (***) | Montant non remboursé Assurance Maladie Obligatoire | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 46 | HBLD158 | Couronne Céramo-céramique | 3 | 2 | 550,00 | 550,00 | 120,00 | 84,00 | 466,00 | |
| 1 | 1 | 46 | HBLD724 | Couronne provisoire | 2 | 60,00 | 60,00 | 10,00 | 7,00 | 53,00 | ||
| 2 | 2 | 47 | HBMD460 | Onlay Céramique dentaire NF EN ISO 6872 | 3 | 3 | Aucun | 450,00 | 100,00 | 70,00 | 380,00 | |
| 3 | TOTAL € (des | actes | envisagés) | 1 060,00 | 230,00 | 161,00 | 899,00 |
And there we go! You can now process the dataframe using whatever business logic applies to your problem. Hopefully this article might have saved you some previous minutes.
Amazon Textract is very good if you’re processing a lot of files that have the same template. Alas, at my job, there are many templates, and Textract doesn’t work on each of them. I thus use a different kind of logic where I extract annotations through an OCR and reconstruct rows myself with a clustering algorithm. I have yet to look into other table extraction tools such tabula-py and Camelot.
Stay safe!